User Guide for NeXO Web Application
- Hierarchical layout of the complete ontology according to its most informative parent-child term relations, enabling the entire ontology to be explored as a multi-scale geographic map.
- Semantic zooming, providing details on demand so the user sees different amounts of detail in a view by zooming in and out.
Navigation
Terms are nodes. Each node in the ontology (NeXO or GO) represents a functional group of genes or a “term”. In GO, terms are labeled with the cellular component, process, or function they represent. In NeXO, terms are labeled based on the best alignment of the data-driven ontology to the GO Cellular Component ontology.
Term relations are edges. Each edge in the ontology (NeXO or GO) represents a parent-child relation between terms. Such relations can have either of two meanings:
- The child term is a part of the parent term (part_of relation).
- The child term is a type of the parent term (is_a relation).
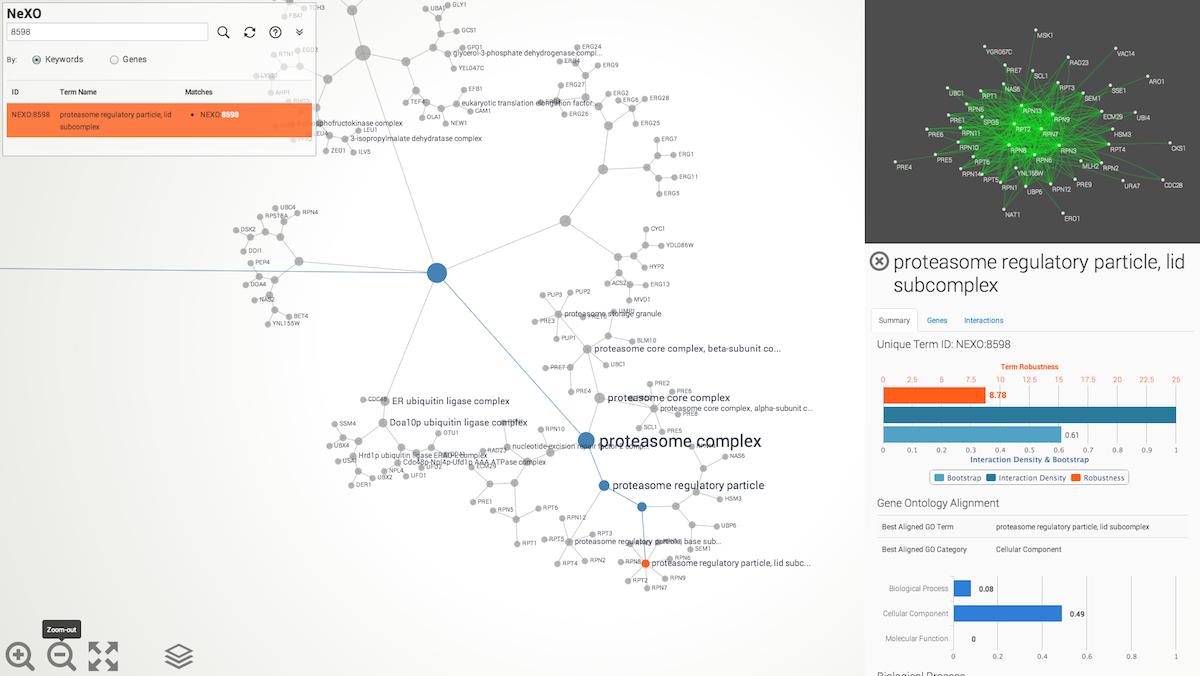
Browsing. Browse the NeXO or GO ontology using the mouse. Click-and-drag to pan. Use the mouse wheel (two-finger scroll) to zoom in or out of selected regions of the ontology. Click on an ontology term to select it. When a term is selected, the relations to ancestral terms are highlighted and a term summary view is presented with information about the selected term (see below). Double-click on the page background to reset the current selection and re-zoom the ontology to fit the page.
Basic navigation buttons
Use the navigation buttons (lower left) to zoom-in and out of the ontology, fit the ontology layout to screen and select the ontology to be visualized (NeXO or GO). Zoom-in
Zoom-in Zoom-out
Zoom-out Fit to Screen
Fit to Screen Ontology selector: choose which ontology to
visualize
Ontology selector: choose which ontology to
visualizeSearch
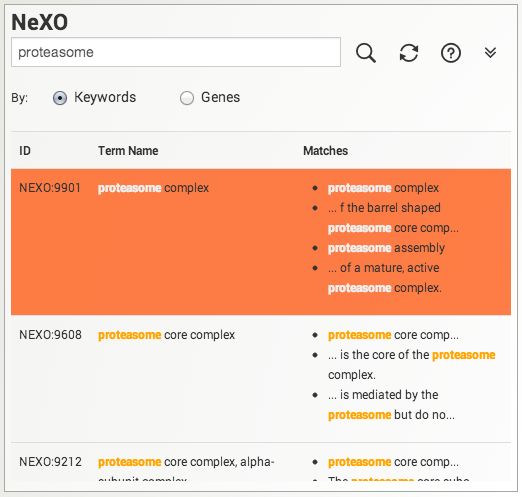
Searching for terms or genes. Search the NeXO and GO ontologies either by keyword or by gene. Results are displayed below the search box. Click on a search result to select and highlight a gene or term in the displayed ontology. Use the refresh button to clear search results and the search box.
When performing searches please keep in mind the following:
- Each search result must contain all words in the query.
- Multiple words encased in double quotes are treated as a single phrase.
- Queries are case insensitive
Displaying term summary information
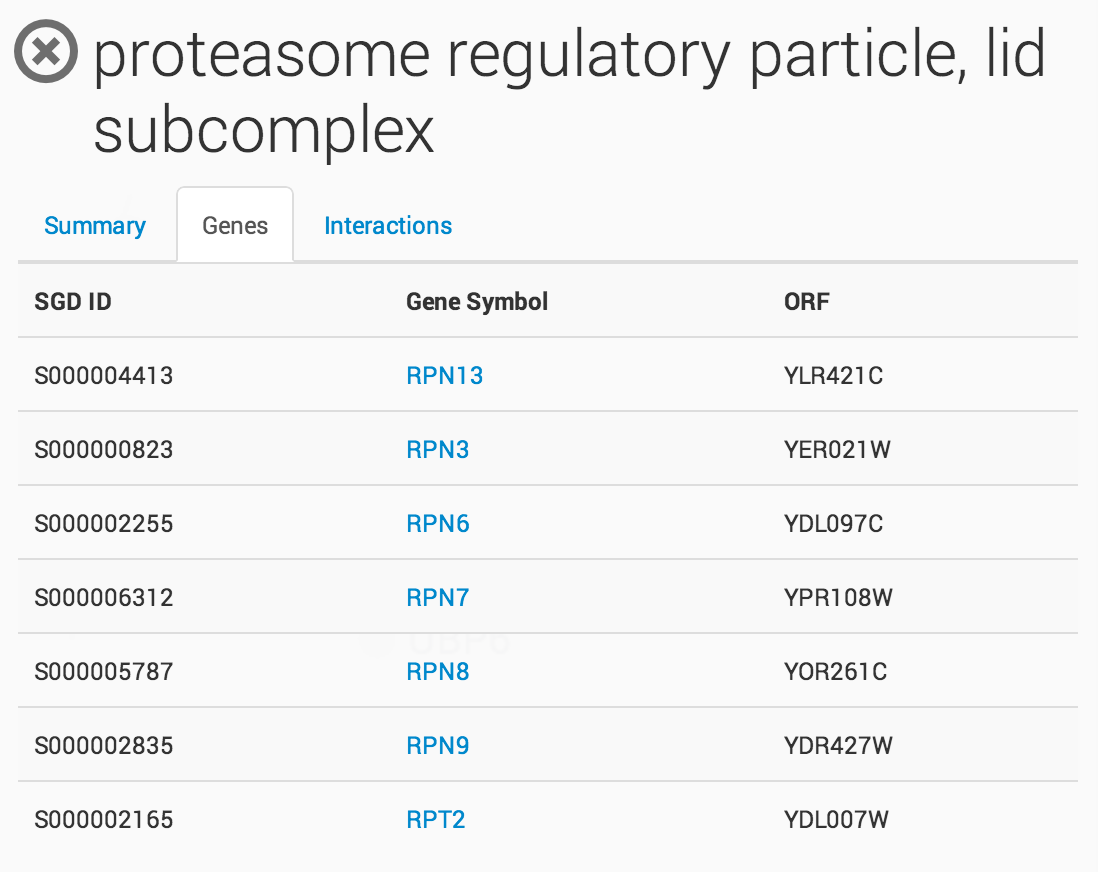
Clicking on a term in the ontology opens a summary information panel for the selected term. This information panel includes the term name, description and associated genes. For NeXO terms, the information panel also includes statistics about the support for the term in network data (bootstrap score, term robustness and interaction density), the alignment of the term to each of the branches of the Gene Ontology and a list of interactions among the genes annotated to the term.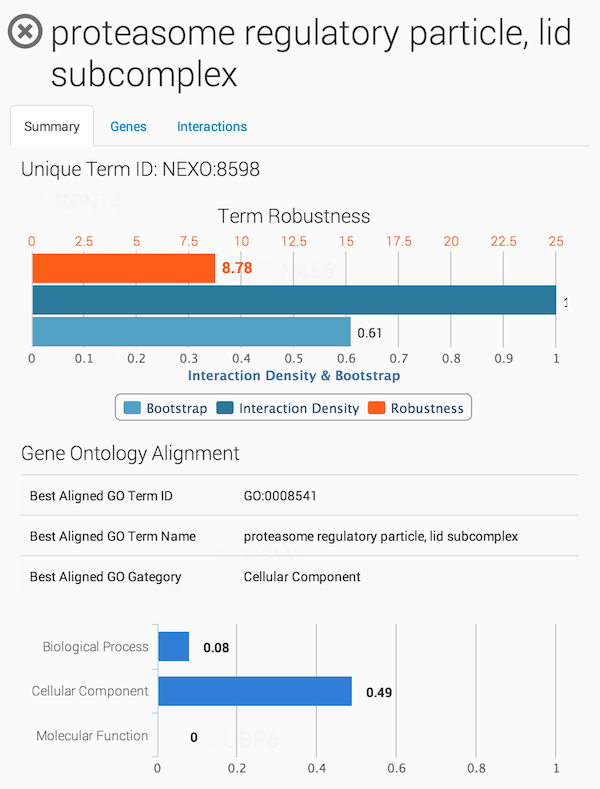
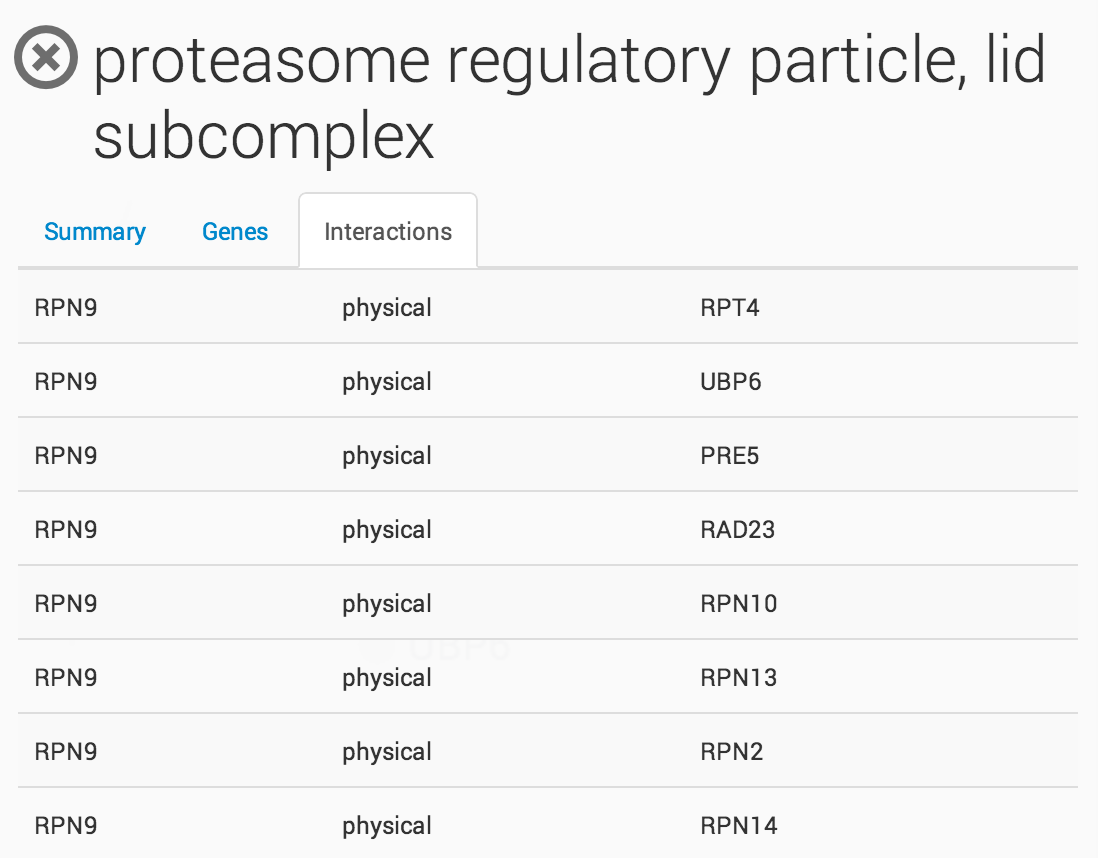
Raw Interaction view
Term Enrichment
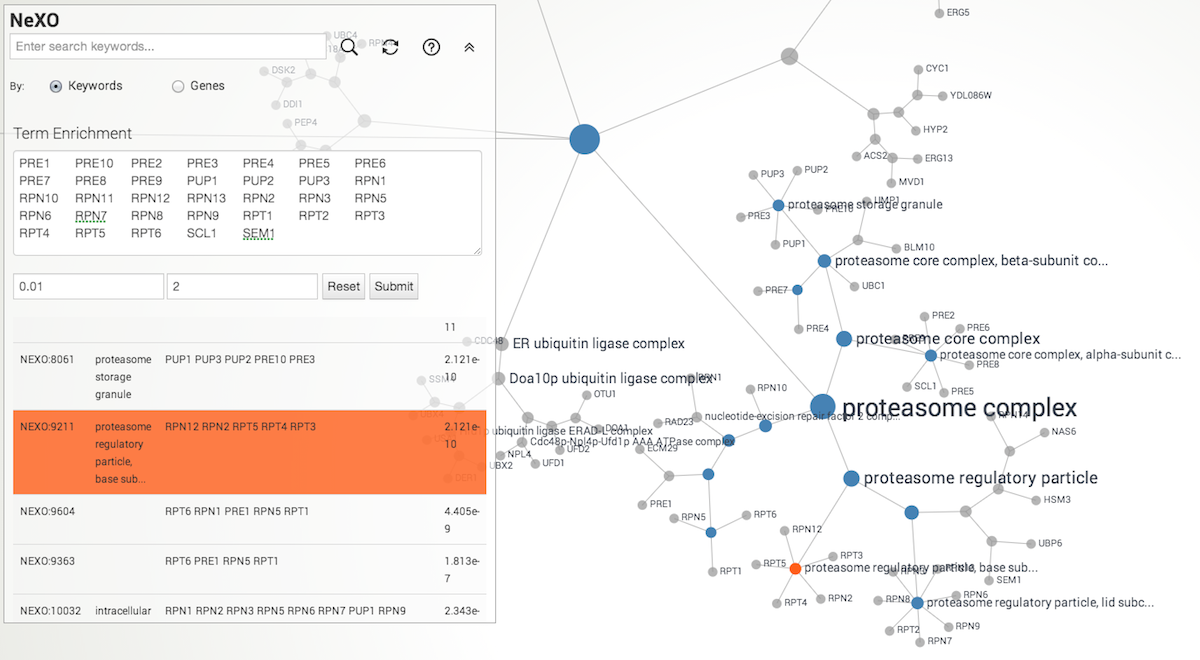
NeXO web app supports term enrichment for yeast genes. Simply enter list of genes of your interest in the text box. Default p-value cutoff is 0.01, and minimum number of genes assigned to the term is 2. Currently, SGD ID, ORF name, and gene symbol are supported as input.